For installing R packages, CRAN is the main place to go. It’s a fantastic free service that checks R packages to make sure they meet standards and then compiles them for multiple architectures. It’s largely a volunteer effort and is important to keep packages working together. Bioconductor is a more specialized repository. Getting a package to pass the checks to go on CRAN has long been a point of pride, and it does lead to higher quality packages, though with odd constraints (the overall package size must be small, all examples must run very quickly, and more).
However, keeping a package on CRAN is an issue. Requirements keep changing: will this package work on the development version of R on Debian with a clang compiler, Fedora with a gcc compiler, Mac with both arm and x86 chips, Windows, etc. and continue to do so as operating systems, hardware, and compilers change? For example, our hisse package was removed briefly from CRAN in spring 2023 (it’s back online) because of an issue where, while the package passed all checks for Mac, Windows, Linux, all R-devel, R-patched, R-release, and R-oldrel, it would generate a warning if compiled using the -Wlto-type-mismatch flag to GCC. Our code hadn’t changed, and this wouldn’t have affected any users, but CRAN gave us two weeks to fix the warning. We didn’t do it on time, so it was archived, which does affect users (making it impossible for most users on Mac or Windows to use it, for example, as they don’t have compilers installed).
If you are part of an ecosystem, there are further risks. Many phylogenetics packages depend or import geiger – CRAN removed that package, and then two weeks later would have also archived every package that depends on it. Geiger’s authors got it restored in time, but it shows how brittle CRAN’s policies make our community. It’s like the Randall Munroe XKCD cartoon:
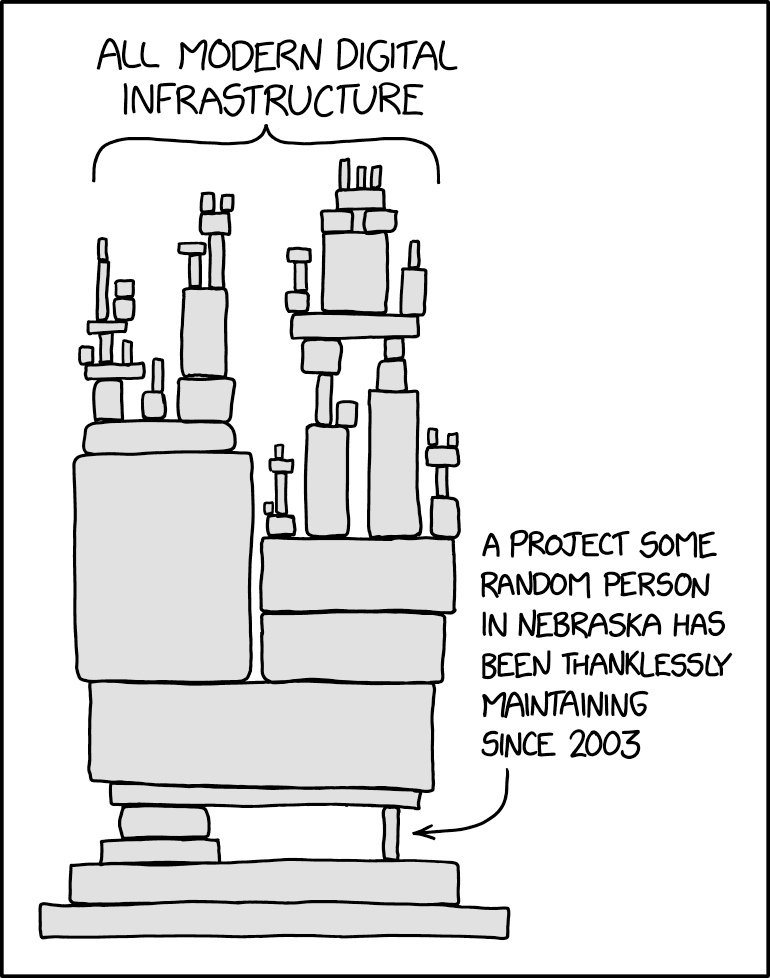
but with the poor maintainer in Nebraska putting the whole ecosystem at risk if they take two weeks off and there is a new requirement in the interim.
The nature of academia makes this even more precarious. A student might work on a package for their dissertation and others could find it very useful. There is no guarantee that the student remains in the field: it’s not unusual for someone to go into teaching, or working at a nonprofit, or working for a company, and even the weak incentive of getting more citations is no longer sufficiently relevant for them to volunteer their time to meet changing CRAN standards. Ongoing maintenance would be ideal for the field, but it is a high bar.
Fortunately, rOpenSci has made an alternative: r-universe. This creates a place where R packages can be built automatically for different platforms, just as CRAN does, but in nearly real time. If a package is in an open source repository (the best known is GitHub), and the r-universe instance knows about it, it will try to compile it. Thus, even if something goes off CRAN, students taking a class can likely still install it from r-universe.
To help our community, I’ve created an instance for phylogenetics. The instance is at https://phylotastic.r-universe.dev/ . Information on how to use it is at http://phylotastic.org/phylotastic.r-universe.dev/. For the packages, I’ve used everything from the CRAN task view for phylogenetics (something I maintained for over a decade, now taken over by Dr. William Gearty who has assembled a team to keep it going), as well as other packages used in phylogenetics in R, including packages in development.
Using the universe
To use the most recent available version of each package:
options(repos = c(
phylotastic = 'https://phylotastic.r-universe.dev',
CRAN = 'https://cloud.r-project.org')
)This will check the phylotastic r-universe, and only go to CRAN for packages that are not there.
To use the CRAN package if available, then use phylotastic as a backup:
options(repos = c(
CRAN = 'https://cloud.r-project.org',
phylotastic = 'https://phylotastic.r-universe.dev')
)Why “phylotastic”?
Phylotastic started as an idea launched at the [US] National Evolutionary Synthesis Center, NESCent, in 2012. Over hackathons and other events, and later an NSF grant (written by Arlin Stoltzfus, Enrico Pontelli, and me) it resulted in various tools for phylogenetics (includng http://datelife.opentreeoflife.org/ ). I wanted to keep that connection, not least so that people could use phylotastic R packages even when there are CRAN hiccups.
To subscribe, go to https://brianomeara.info/blog.xml in an RSS reader.
Citation
@online{o'meara2023,
author = {O’Meara, Brian},
title = {An Alternative to {CRAN} for Phylogenetics},
date = {2023-04-23},
url = {https://brianomeara.info/posts/phylotasticruniverse/},
langid = {en}
}